- Biotechnology group>
- NAIST Collaborative Laboratory

| Professor | Masayuki Inui |
|---|---|
| mmg-lab@rite.or.jp |

| D1 | Yurina Onoue |
|---|---|
| M2 | Naoki Amemiya |
| M2 | Fuka Sawada |
| M2 | Yukino Terazawa |
| M2 | Yukitaka Hyodo |
| Visiting Professor | Masayuki Inui |
| Visiting Associate Professor | Takahisa Kogure |
| M1 | Minori Omura |
| M1 | Yosuke Suzuki |
| M1 | Rinno Mine |
| M1 | Tomoka Okuda |
Global warming induced by carbon dioxide and exhaustion of fossil fuels has been considered a big social issues these days. These result from cross-border issues such as energy consumption by developed countries and economic growth of developing countries. To resolve these issues, not only just technology development but also comprehensive understanding of global production and consumption system are required. Molecular Microbiology and Genetics laboratory are focusing on bioprocess by microorganisms using biomass as a renewable resource. We are dealing with technological development using renewable resources which enables circulating and low carbon society.
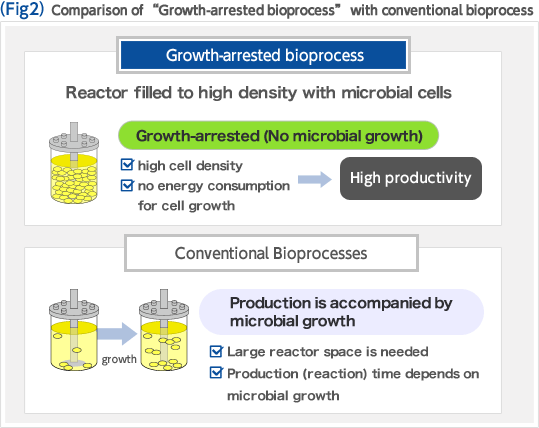
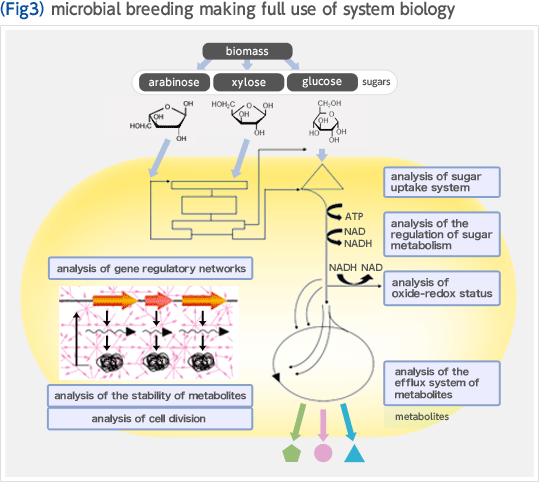
Biorefinery concept is a new concept in which renewable resources such as biomass are converted to fuel and chemicals. In the U.S., where this concept was developed, the government takes initiative in replacement of oil refinery, processing crude oil, with biorefinery (Fig. 1). The development of biorefinery technology is anticipated to play an important role in establishing a sustainable society in the 21st century. We have developed an efficient biomass utilization technology "Growth-arrested bioprocess" based on Corynebacterium glutamicum which has been widely used for industrial production of amino acids. The key to achieving high efficiency and high productivity is the effective separation of the microbial growth phase from the production phase of the target compound. This manner of using bacterial cells as if they were simple chemical catalysts enables one to produce large amounts of chemicals in short periods of time, and unlike conventional bioprocesses, the productivities reached, expressed as space-time-yield (STY), are comparable to those of chemical processes (Fig. 2). We are constantly expanding the range of product options that the growth-arrested bioprocess can support. To this end, we implement global analysis tools including system biology based on metabolome analysis, metabolic pathway design, and genome engineering based on the genome database of C. glutamicum. We are making full use of these methods to optimize production strains (Fig. 3).
We are establishing a fundamental technology using the growth-arrested bioprocess which produces bioethanol from non-food agricultural feedstock such as rice straw and corn stover. We are collaborating with NREL (National Renewable Energy Laboratory) founded by U.S. Department of Energy to achieve actualization of an economically efficient production process for ethanol using non-food biomass. We are also working on the production of biobutanol which is one of expected materials for biojet fuels and green chemicals such as organic acids which can be polymer materials, alcohol, and aromatic compounds.
Development of bioprocess based on metabolic engineering requires not only introduction of biosynthesis pathway and/or expression enhancement of promoter activities, but also optimal adaptation to changes of intracellular and extracellular environment such as engineering of redox balance and improving tolerance against fermentation inhibitors. Therefore, comprehensive understanding of several stress responses (oxidative, acid, alkaline, heat shock, osmotic stress etc.) and regulations of carbon catabolism such as carbohydrate utilization, energy metabolism and cofactor regeneration are subject to intensive studies. We are analyzing functions and regulations of several metabolic- and cell division-related genes based on C. glutamicum whole genome sequencing performed by us. We are studying mechanisms of gene expressions and regulations of regulatory elements. Comprehensive analysis of regulatory elements in C. glutamicum has been revealing unique gene regulatory networks in this bacterium. The understanding of the genome-wide regulatory networks is indispensable for basic information to develop a technology to optimize gene expressions by metabolic engineering.

| Completion | Name | Title |
|---|---|---|
| 2019.3 (Master) | Masato Sawa | Regulation of RNase III expression in Corynebacterium glutamicum. |
| 2019.3 (Master) | Hiroaki Takamitsu | The study on the promoters specific for ECF sigma factors in Corynebacterium glutamicum. |
| 2019.3 (Master) | Hiroki Machida | In vivo screening method for isoprenoid biosynthetic enzymes using Corynebacterium glutamicum. |
| 2018.3 (Master) | Kentaro Nagoshi | Metabolic engineering of Corynebacterium glutamicum for production of methionine. |
| 2018.3 (Master) | Akihiro Domon | Functional analysis of asparagine and aspartic acid metabolism-related genes in Corynebacterium glutamicum. |
| 2018.3 (Master) | Ryoji Ogura | Aptitude analysis of microorganism for producing aromatic compounds by evaluating stress tolerance. |
| 2017.3 (Ph.D.) | Takayuki Kuge | Engineering of Corynebacterium glutamicum for arabinoxylan utilization. |
| 2017.3 (Master) | Yuji Yamamoto | Regulation of the expression of rho in Corynebacterium glutamicum. |
| 2017.3 (Master) | Junya Maeda | Characterization of phenol 2-monooxygenase from Corynebacterium glutamicum. |
| 2017.3 (Master) | Yuta Tsukada | Functional analysis of glucokinase genes in Corynebacterium glutamicum. |
| 2016.3 (Ph.D.) | Toshihiro Tsujimoto | Analysis of the isobutanol stress response in Corynebacterium glutamicum. |
| 2016.3 (Master) | Yuki Tanaka | Transcriptional analysis of the genes involved in trehalose biosynthesis in Corynebacterium glutamicum. |
| 2016.3 (Master) | Yuta Sugimoto | Functional analysis of the RNase Z family enzyme in Corynebacterium glutamicum. |
| 2015.3 (Master) | Nagisa Hamamoto | Analysis of RNase J in Corynebacterium glutamicum. |
| 2015.3 (Master) | Kumiko Tanaka | Transcriptional regulation of the genes involved in the synthesis of aromatic compounds in Corynebacterium glutamicum. |
| 2014.3 (Master) | Takayuki Kuge | Transcriptional regulation of L-arabinose utilization genes in Corynebacterium glutamicum. (Yano Award) |
| 2014.3 (Master) | Hirotaka Hara | Analysis of Translesion Synthesis in Corynebacterium glutamicum. |
| 2014.4 (Master) | Ryosuke Nariai | Analysis of the regulation of prsA gene expression in Corynebacterium glutamicum. |
| 2013.3 (Master) | Toshihiro Tsujimoto | Isobutanol stress response in Corynebacterium glutamicum. |
| 2013.3 (Master) | Terukazu Ito | Genome-wide analysis of the role of global transcriptional regulator GntR1 in Corynebacterium glutamicum. |
| 2011.3 (Master) | Yusuke Oba | Screening of Corynebacterium glutamicum and giving of new carbon source to RITE strain. |
| 2010.3 (Master) | Tomoo Sakamoto | Isolation and characterization of microorganisms for syngas fermentation. |
| 2010.3 (Master) | Toshihiko Iwami | Biohydrogen production - Hybrid-hydrogen production system using Escherichia coli carrying hydA gene and phaC deficient strain of Rhodobacter sphaeroides. |
| 2009.3 (Master) | Yumi Natsuma | Studies on valine production by the RITE bioprocess. |
| 2009.3 (Master) | Eiji Mori | Metabolome analysis for efficient alanine production in Corynebacterium glutamicum. |
| 2008.3 (Master) | Akihumi Omori | Microbial ecology of a deep subsurface ecosystem and investigation of CO2 fixation genes. (Best Student Award of NAIST) |
| 2008.3 (Master) | Atsushi Ota | Construction of an efficient hydrogen producing strain by heterologous expression of hydrogenase gene, hydA. |
| 2007.3 (Ph.D.) | Youta Tsuge | Random genome deletion studies of Corynebacterium glutamicum. (Research Fellowship for Young Scientists from JSPS) |
| 2007.3 (Master) | Takeshi Eimoto | H2 production by water-gas shift reaction. |
| 2007.3 (Master) | Takashi Imamiya | Transcriptional regulation analysis of Corynebacterium glutamicum methionine biosynthesis genes in RITE process conditions. |
| 2006.3 (Master) | Shou Ishii | Culture independent analysis of microbial diversity and cloning of CO2 fixation gene in deep subsurface. (Best Student Award of NAIST) |
| 2005.3 (Master) | Daisuke Hanayama | Production of ethanol from biomass hydrolysates by genetically engineered coryneform bacteria. |
| 2005.3 (Master) | Satoshi Okayama | Establishment of genome engineering in Corynebacterium glutamicum. |
| 2002.3 (Master) | Asako Yamaguchi | Production of porphyring by genetically engineered coryneform bacteria. |
-

HPLC
-

Anaerobic Chamber
-

Electronic Mashing Device for saccharification of biomass
-

Thermal cycler
-

Typhoon Image Analyzer
-

Jar fermentor
-

LC-MS/MS
-

GC
-

DNA microarray scanner
-

chromatography system
-

electron microscope
-

confocal microscope
-

Robotic workstation for automated purification of plasmid

![[3]Analysis of gene functions and regulation of gene expression in C. glutamicum](https://www.rite.or.jp/bio/en/naist/img/tit05.gif)